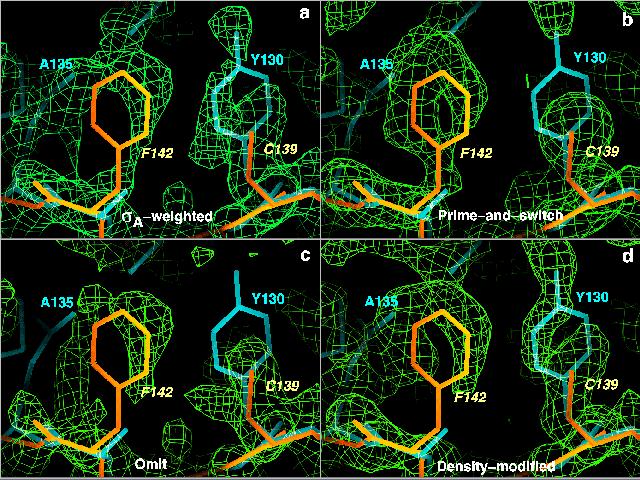
X-ray data on P. aerophilum IF5A to 2.2 A (Peat T.S., Newman J, Waldo G.S, Berendzen J. & Terwilliger T.C. Structure Of Translation Initiation-Factor 5a From Pyrobaculum aerophilum At 1.75 Angstrom Resolution. Structure 6, 1207-1214 (1998))
60% solvent
atomic model from M. jannaschii IF5A (Kim, K. K, Hung, L. W., Yokota, H., Kim, R. & Kim, S.-H. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 angstrom resolution. Proc. Natl. Acad Sci. USA 95, 10419-10424 (1998))
RMSD between atomic model from M. jannaschii and refined P. aerophilum structure is 1.7 A.
The test: phase the P. aerophilum data with the M. jannaschii structure
Methods tested: (a) sigma-A, (b) prime-and-switch, (c) omit map, and (d) density modification (with RESOLVE)
Results: (MR model in blue, correct model in yellow). Sigma-A map looks a lot like the model in blue used to calculate it; prime-and-switch map looks just like the correct model in yellow. Omit map is noisy, and density-modified map is halfway between sigmaA and prime-and-switch.

![]()
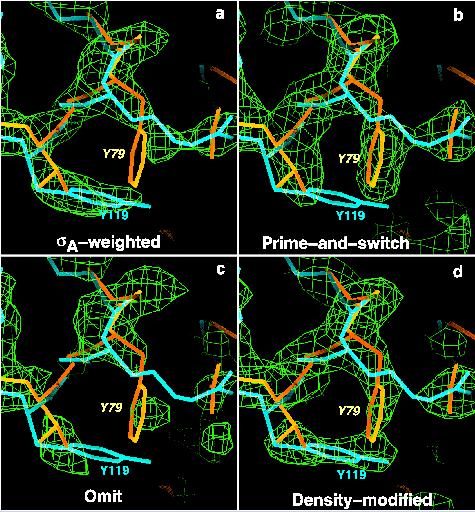
The system: Rhodococcus dehalogenase (dhlA)
X-ray data on Rhodococcus dhlA to 1.5 A (Newman, J., Peat, T. S., Richard, R., Kan, L., Swanson, P. E., Affholter, J. A., Holmes, I. H., Schindler, J. F., Unkefer, C. J., & Terwilliger, T. C. Haloalkane dehalogenases: Structure of a Rhodococcus enzyme. Biochemistry 38, 16105-16114 (1999).)
30% solvent
atomic model from dehalogenase linB from S. paucimobilis(Marek, J., Vevodova, J., Smatanova, I. K., Nagata, Y., Svensson, L. A., Newman, J., Takagi, M. & Damborsky, J. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39, 14082-14086 (2000).)
RMSD between atomic model from S. paucimobilis lin B and refined RhodococcusdhlA structure is 1.4 A.
The test: phase the Rhodococcus dhlA data with the S. paucimobilis linB structure
Methods tested: (a) sigma-A, (b) prime-and-switch, (c) omit map, and (d) density modification (with RESOLVE)
Results: (MR model structure is in blue, correct model in yellow). SigmaA map looks like the MR model used to calculate it; prime-and-switch map looks like the correct model.